Exploring the Movement Database
After week 1, the new priority was to find the right data on the internet. I started the week off by reading papers based on locomotion in the C. elegans worm. Surprisingly enough (or unsurprisingly, depending on how much you’re into worms), most of the data was from the OpenWorm Movement Database.
Almost all the video data on the database is from the Tierpsy tracker, which is an amazing worm tracking tool in itself. But there was a new hurdle now.

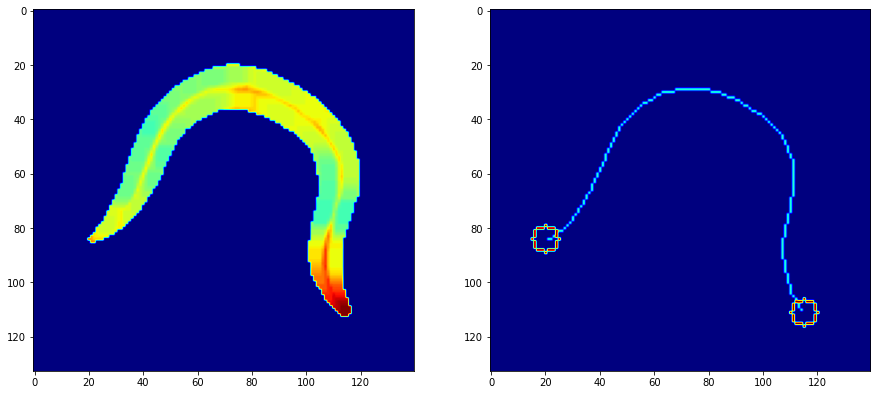
The issue with the video data (as seen above) wThe issue with the video data (as seen above) was that the worms were “partially segmented out”. Which meant that the background was all blacked out except for a small “padding area” around the worm. This “padding area” was a good safety measure on the Tierpsy tracker’s side, but was a minor hurdle for someone who wanted to segment out just the worm from the image in order to gain inferences from behavioural dynamics.
This hurdle was taken care of in 3 stages:

-
Stage 1: Extracting the “padding region” around the worm with
cv2.threshold() -
Stage 2: Removing the padding region by using
cv2.bitwise_and()between the real and the thresholded image -
Removing any extra noise (as seen on stage 2) with
cv2.erode()
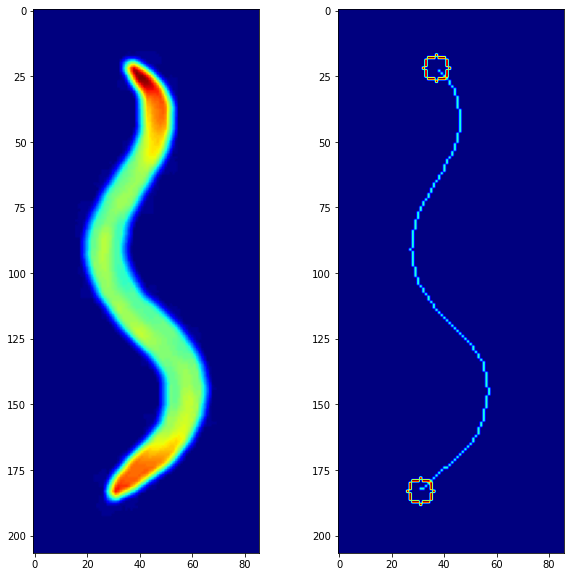
Now the next objecive was to extract the head and tail, this was done using corner detection with cv2.goodFeaturesToTrack() with maxCorners = 2 to only capture the 2 most prominent corners.
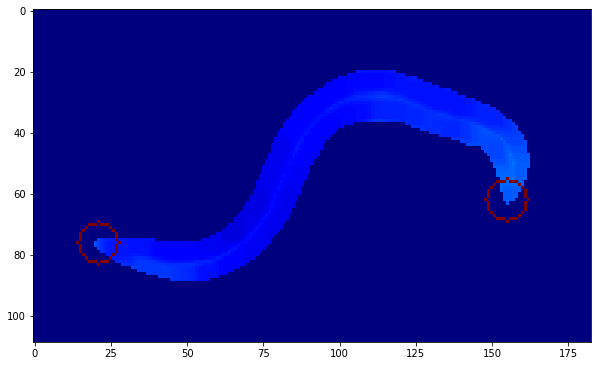
To complete the skeletonization, all that was needed now was to skeletonize the main body, and that was done with the very self explanatory function named skimage.morphology.skeletonize(). Combined with two circles which signify the head and the tail, we get a clean skeleton which can now be used for analysis on a bigger scale with a large number of frames.