DevoLearn (Open-source) Maintenance and Evangelism
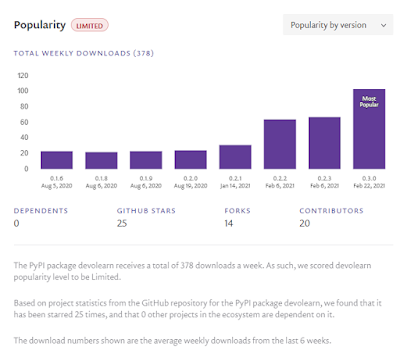
2021 has been a busy year for the DevoLearn initiative. Not only has Mayukh Deb been busy maintaining and generating new versions of the DevoLearn pre-trained model, but I (Bradly Alicea) has been engaging in technology evangelism to advance awareness and involvement in the initiative. The DevoLearn pre-trained model software (for C. elegans embryogenesis) is now at version 0.3.0, and has garnered 12 contributors making 165 commits (largely since January 2021). Our involvement in Google Summer of Code has bolstered many of these contributions. While our popularity is currently limited, we are trying to spread the word.
To that end, we have presented two versions of a promotional talk on using DevoLearn for facilitating Computational Developmental Biology research and education. The first presentation (DevoLearn: Engaging learners with Computational Developmental Biology) is a flash talk given to the OSF Education Un-conference on Open Scholarship Practices in Education Research, held in February. The second presentation was a longer (15-minute) presentation to the INCF Assembly (DevoLearn: a platform for open Developmental Data Science, Machine Learning, and Education), held in April.

As for developing the broader platform, Ujjwal Singh and I will be working this Summer to develop algorithms for colony morphogenesis and behavior in the Diatom genus Bacillaria. This will be added to the platform in a manner similar to the DevoLearn pre-trained model. In addition to the software development activities, Mayukh, myself, and Krishna Katyal have been the main contributors to the DevoWorm Onboarding Guide. Looking forward to an exciting future!
Update, 8/19:
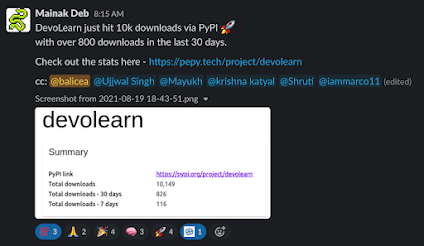
All of this work has paid off! From the #devolearn Slack channel (OpenWorm Slack team).
Posted by Bradly Alicea