Upcoming Update on DevoWorm Project to OpenWorm

Next Friday (4/15) at 9:00am Pacific Time, I will be presenting an update to the OpenWorm Journal Club on advances in the DevoWorm subproject (How a Worm Develops). It has been a year and a half since the previous update [1], and we have made significant progress on a number of fronts:
-
as of right now, our group consists of myself, Richard Gordon, Tom Portegys, Steve McGrew, and Gabriel Pascualy.
-
DevoWorm now consists of three interests groups, all of which are fairly informal: Digital Morphogenesis, Developmental Dynamics, and Reproduction and Developmental Plasticity. It is hoped that as the project matures and attracts more collaborators, the interest groups will keep the subproject focused on specific goals.
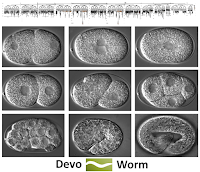
Tom, Steve, and Gabriel have been taking the lead on the Morphozoic platform, which is part of the Digital Morphogenesis interest group. Morphozoic is a hybrid model (Cellular Automata/ANN) that can approximate morphogenetic processes. The Cellular Automata component utilizes an approach called nested neighborhoods that captures the action of cell-cell communication and signaling gradients in a way conventional Moore neighborhoods do not. Tom has also produced a number of demos ranging from simulating biological pattern formation to image processing. This work will be featured in an soon to be published book chapter [2].
Richard Gordon and myself have been taking the lead on the Developmental Dynamics interest group. To this end, we have worked out differentiation trees [3] for Caenorhabditis elegans [4] and Ciona intestinalis [5]. Differentiation trees are essentially reorganizations of the lineage tree based on the size differential of daughter cells after a cell division event, and may point us to subtle spatial patterns such as the precursors of tissue formation. More generally, we have been attempting to work out cross-species comparisons of early embryonic development, as well as novel computational characterizations of both mosaic and regulative development in multiple species. Some of this work will be featured in an upcoming publication in a special issue of the journal Biology [6].
The Reproduction and Developmental Plasticity interest group is focused on the evolution and development of C. elegans life-history, and stems from work I did in Nathan Schroeder’s Laboratory at UIUC [7, 8]. So far, this interest group has involved experimental evolution and the induction of developmental plasticity resulting from L1 larval arrest in mutant genotypes. This is the newest area of DevoWorm, but is a necessary component of understanding for working towards whole-organism simulation.

All three DevoWorm project interest groups in their "2-cell phenotype".
If you are interested in joining the DevoWorm group or just attending one of our group meetings, please attend the OpenWorm presentation or contact one of the current group members. More generally, the OpenWorm project is currently recruiting volunteers, so fill out an application and state your skills and specific interests. We are looking for people with a diversity of backgrounds, from hard-core programming and data analysis skills to science communication specialists and biologists with an interest in theoretical synthesis.
NOTES:
[1] Alicea, B., McGrew, S., Gordon, R., Larson, S., Warrington, T., and Watts, M. (2014). DevoWorm: differentiation waves and computation in C. elegans embryogenesis. bioRxiv, doi:10.1101/009993
[2] Portegys, T., Pascualy, G., Gordon, R., and Alicea, B. (2016). Morphozoic: cellular automata with nested neighborhoods as a novel representation for morphogenesis. Forthcoming in Multi-Agent Based Simulations Applied to Biological and Environmental Systems.
[3] Gordon, R. (1999). The Hierarchical Genome and Differentiation Waves: novel unification of development, genetics and evolution. World Scientific and Imperial College Press, Singapore and London.
[4] Alicea, B. and Gordon, R. (2016). Caenorhabditis elegans Embryonic Differentiation Tree (10 division events). doi:10.6084/m9. figshare.2118049.
[5] Alicea, B. and Gordon, R. (2016). Ciona intestinalis Embryonic Differentiation Tree (1- to 112-cell stage). doi: 10.6084/m9.figshare.2117152.
[6] Alicea, B. and Gordon, R. (2016). Quantifying mosaic development: towards an Evo-Devo Postmodern Synthesis via differentiation trees of embryos. Biology (Special Issue: beyond the modern evolutionary synthesis). Submitted.
[7] Alicea, B. (2016). Evolution in Eggs and Phases: experimental evolution of fecundity and reproductive timing in Caenorhabditis elegans. bioRxiv, doi:10.1101/042143.
[8] Alicea, B. (2016). Genotype-specific developmental plasticity shapes the timing and robustness of reproductive capacity in Caenorhabditis elegans. bioRxiv, doi:10.1101/045609.
Posted by Bradly Alicea