Finding Your Inner Modeler (Part II)
Last year, I attended a workshop at the University of Illinois-Chicago called “Finding Your Inner Modeler”. Sponsored by the NSF, FYIM is meant to bring together biologists and modelers and to foster collaborations between the two. There were many interesting talks over the course of two days, including plant biology, biochemical kinetics, and (of course) various types of computational and statistical model [1].
This year was the second installment of FYIM, and this time I was chosen for a platform presentation. The platform presentation (Process as Connectivity: models of interaction in cellular systems) involves a 40-minute discussion between the principal investigator and an expert modeler. For my talk, this expert modeler was Dr. Eric Deeds from the University of Kansas.

The talk features work with several collaborators, features work from the DevoWorm group. In the talk, I described the DevoWorm group as an example of data science biology [2]. As an affiliate of the OpenWorm Foundation [3], the DevoWorm group works with primary and secondary data, and produces secondary and tertiary open datasets that serve as material for publications, student projects, and the wider development/computational biology communities.
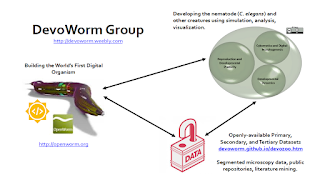
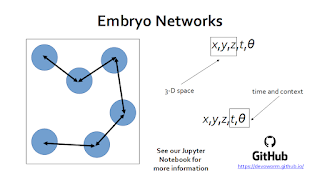
The core innovation introduced in this talk is the use of graph theory and complex networks to analyze the organizational structure of the embryonic phenotype. This work is now showcased in a new paper [4] and Github repository.
NOTES
[1] One example is the Virtual Cell software project, which allows one to model and analyze representations of kinematics, kinetics, geometry, and network interactions at the cellular level.
[2] Alicea, B., Gordon, R., and Portegys, T.E. (2018). DevoWorm: data-theoretical synthesis of C. elegans development. bioRxiv, doi:10.1101/282004.
[3] Sarma, G.P., Lee, C-W., Portegys, T., Ghayoomie, V., Jacobs, T., Alicea, B., Cantarelli, M., Currie, M., Gerkin, R.C., Gingell, S., Gleeson, P., Gordon, R., Hasani, R.M., Idili, G., Khayrulin, S., Lung, D., Palyanov, A., Watts, M., Larson, S.D. (2018). OpenWorm: overview and recent advances in integrative biological simulation of Caenorhabditis elegans. Philosophical Transactions of the Royal Society B, 373, 20170382. doi:10.1098/rstb.2017.0382.
[4] Alicea, B. and Gordon R. (2018). Cell Differentiation Processes as Spatial Networks: identifying four-dimensional structure in embryogenesis. BioSystems, doi:10.1016/j.biosystems.2018.09.009.
Posted by Bradly Alicea